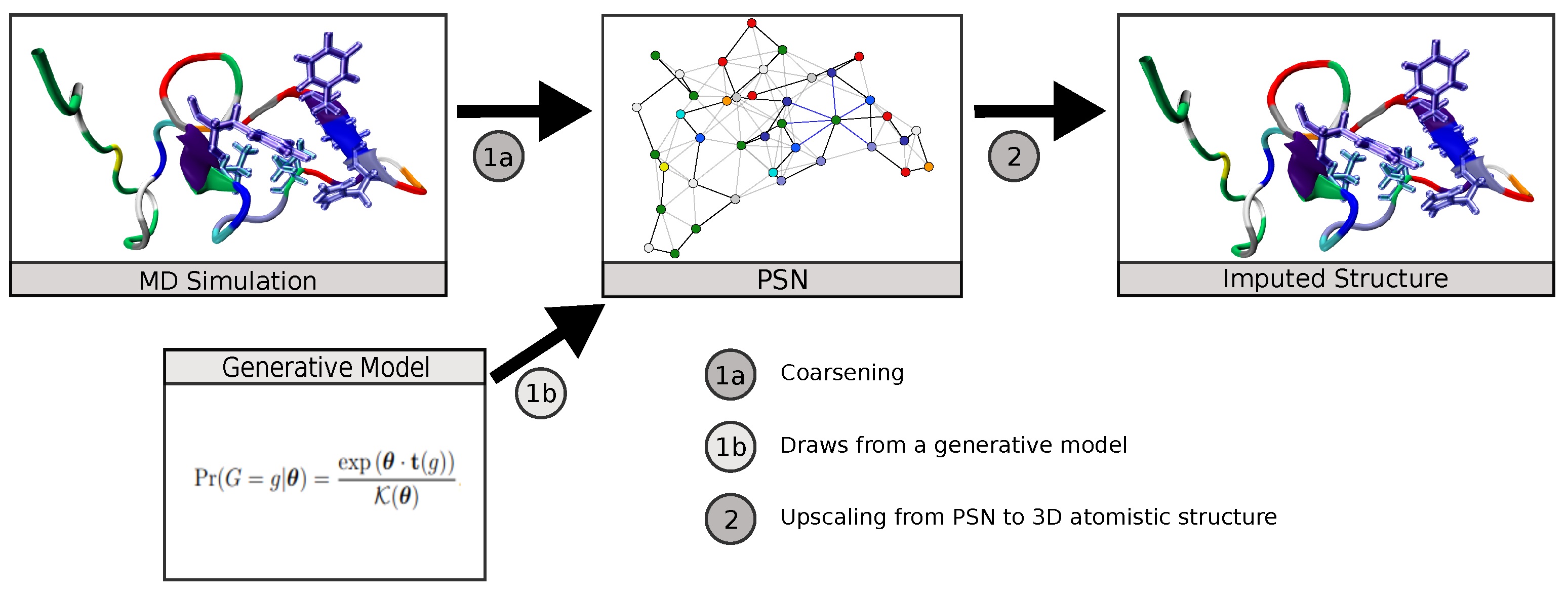
Biomolecules | Free Full-Text | Neural Upscaling from Residue-Level Protein Structure Networks to Atomistic Structures | HTML
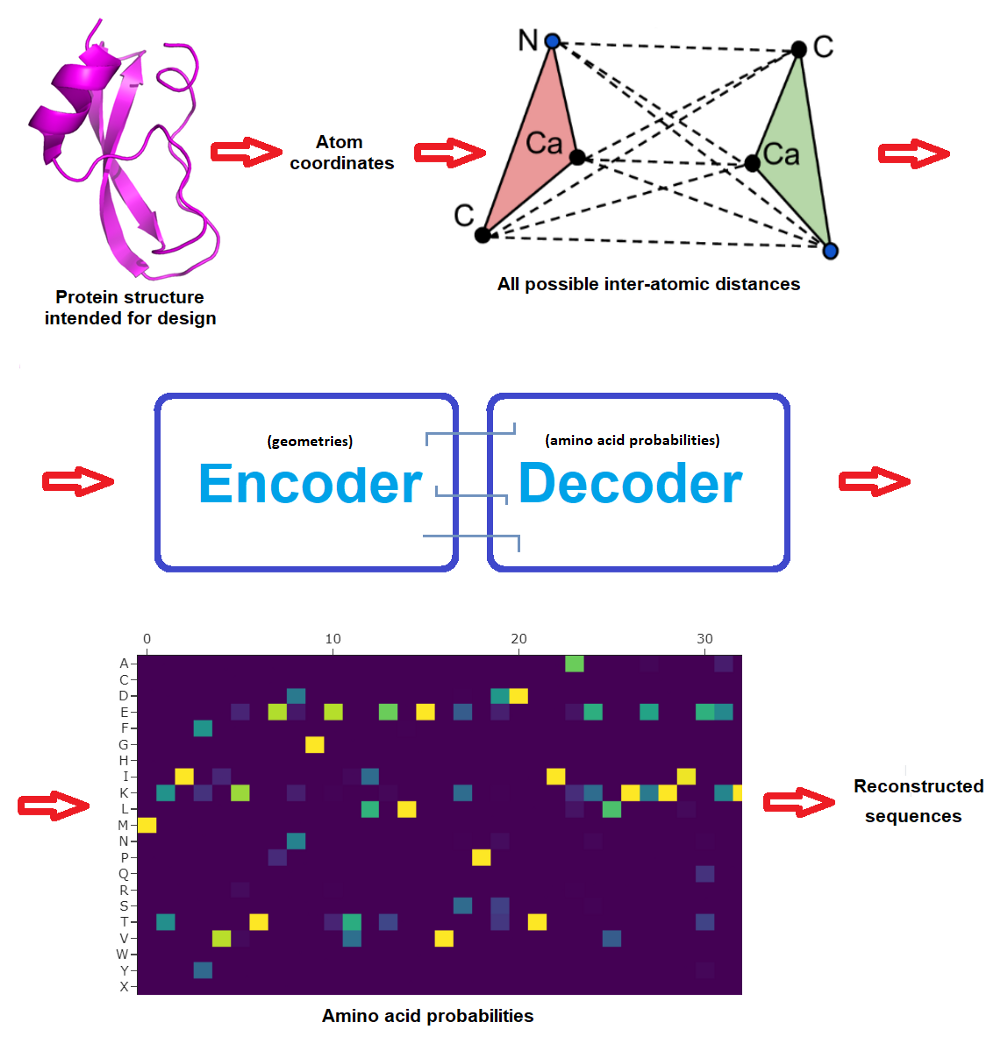
New deep-learned tool designs novel proteins with high accuracy | by LucianoSphere | Towards Data Science

Overview of different machine learning approaches for protein design.... | Download Scientific Diagram
Ig-VAE: Generative modeling of protein structure by direct 3D coordinate generation | PLOS Computational Biology

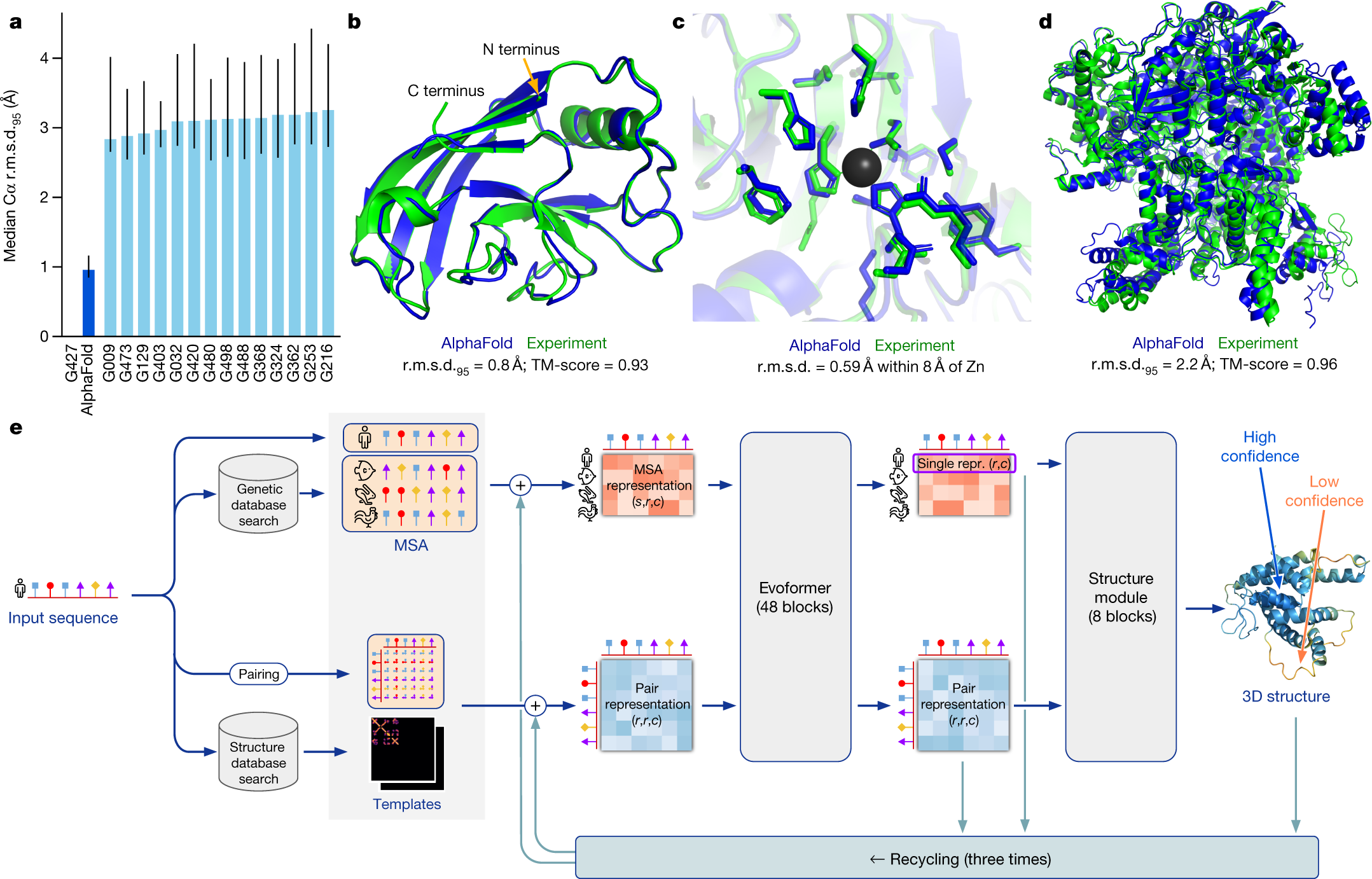
Ultrafast end-to-end protein structure prediction enables high-throughput exploration of uncharacterized proteins | PNAS

From lattice-protein sequence space to inferred Potts model. Protein... | Download Scientific Diagram



![PDF] Generative modeling for protein structures | Semantic Scholar PDF] Generative modeling for protein structures | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/6449a699e79f46c11c6cd0a6797ab7466533a14b/6-Figure3-1.png)
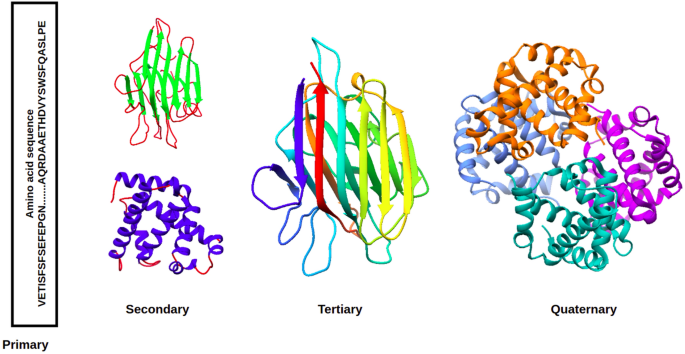
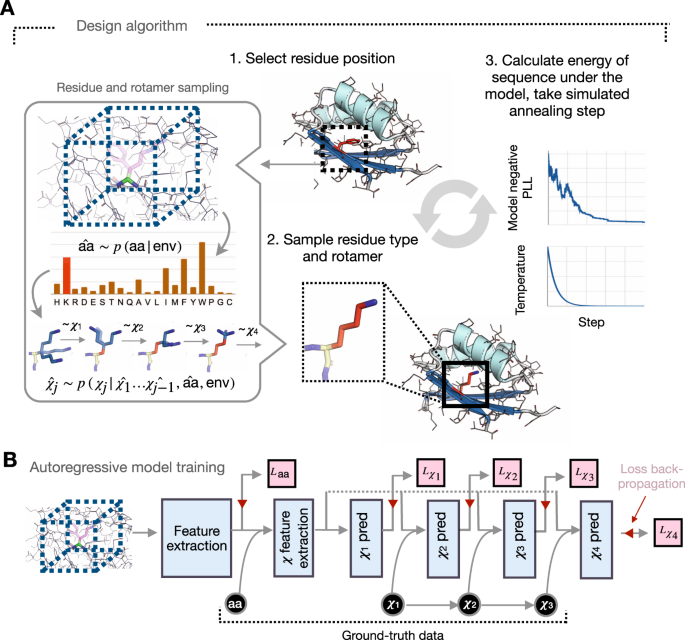
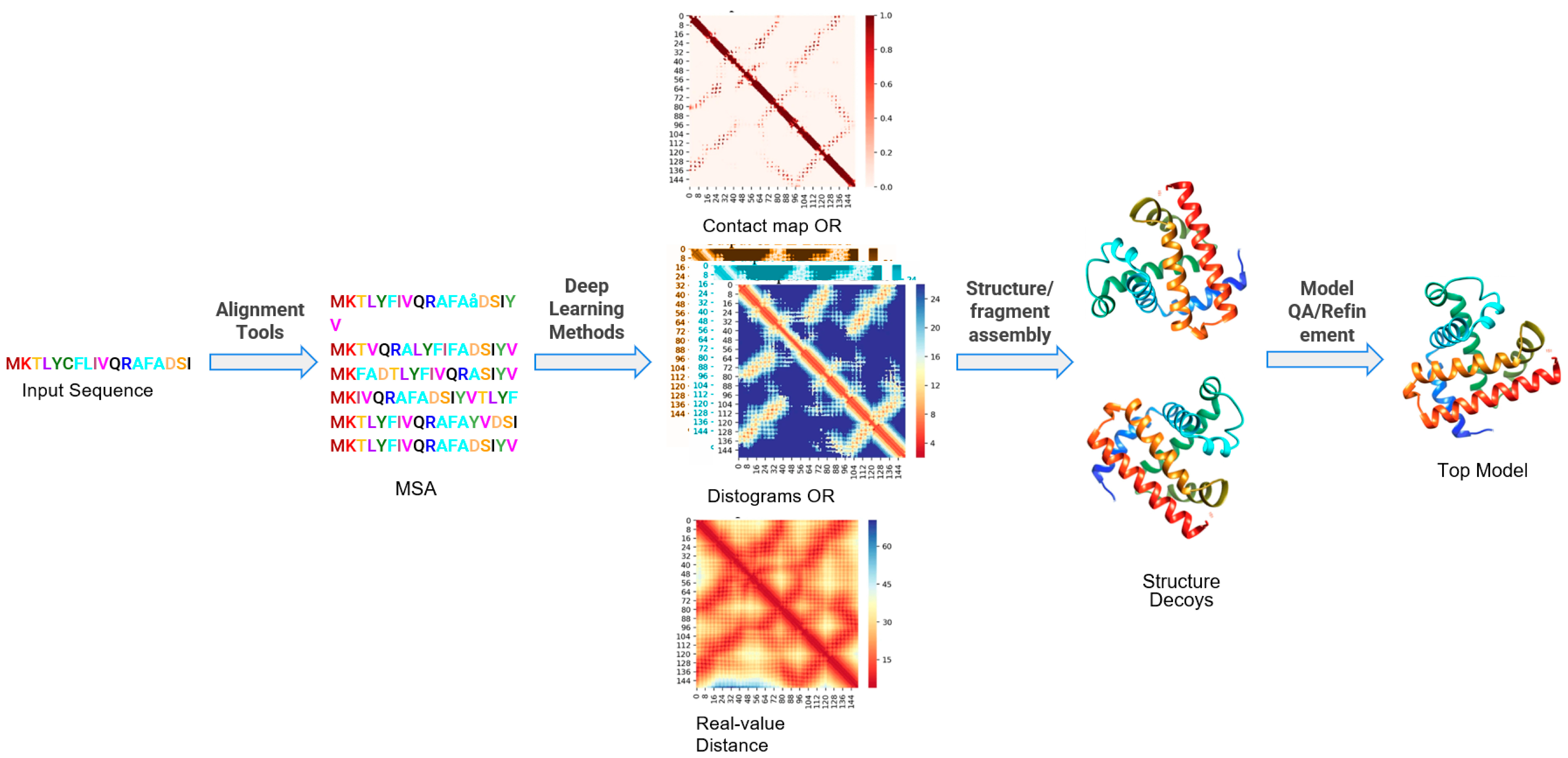
![PDF] Deep Learning in Protein Structural Modeling and Design | Semantic Scholar PDF] Deep Learning in Protein Structural Modeling and Design | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/a08c2b0f7ac943f0cb05e064a477204db4f3fee2/9-Figure3-1.png)